CORDE project
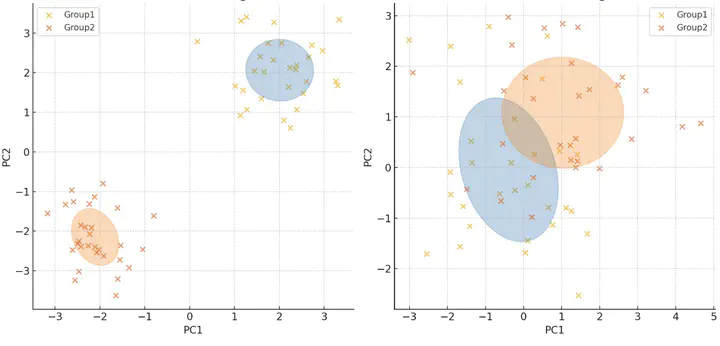 Photo by rawpixel on Unsplash
Photo by rawpixel on UnsplashDimensionality reduction is a central tool in microbiome beta diversity analysis. However, unsupervised methods such as PCoA may fail to reveal relationships between sample clustering and group variables, even when PERMANOVA suggests significant associations. In contrast, supervised approaches like sPLS-DA often overly emphasize the group signal, sacrificing much of the total variation and dissimilarity structure of the microbiome data. Hence, we propose a novel outcome-guided dimensionality reduction framework that balances group-discriminative representation, abundance preservation, and confounder removal. Our approach first performs PCoA on the abundance table and removes confounder-driven components via projection onto the orthogonal complement of the confounder space. We then apply sparse PLS to extract outcome-associated components and compute a taxa-level LDA score. This unified score quantifies each taxon’s contribution to outcome-associated beta diversity, enabling both global comparison and targeted driving taxa identification. The R package is now available at https://github.com/bioscinema/CORDE.